Review of Project DEBBIE - Biomaterials Ontology and Database
Last Updated on February 16, 2023 — 4 min read
This blog series features open-source, ontology-based software solutions that empower scientific research. With the advancements in scientific ontologies, the practical usage in software development is increasingly active but, in my opinion, has yet to reach its full potential. The purpose of this series is to help understand the technical landscape for ontology-based scientific software, highlight valuable use cases and examine the limitations that may impact adoption.
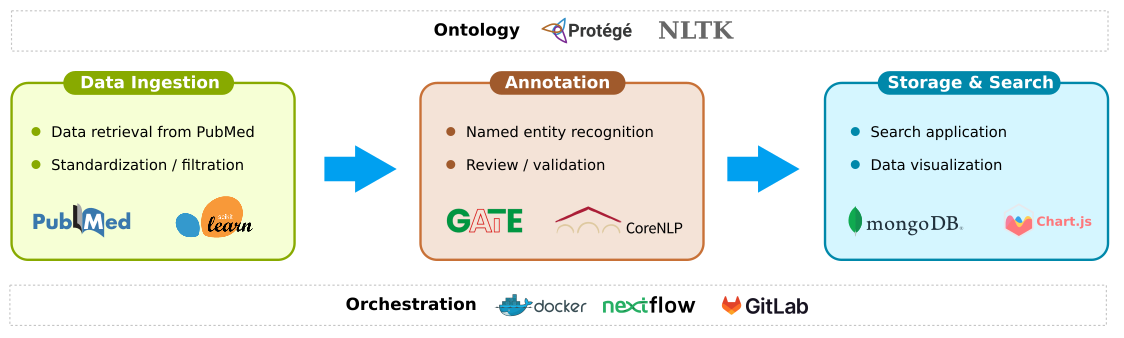
This article focuses on DEBBIE (Database of Experimental Biomaterials and their Biological Effect), a solution that emerged from an EU-funded research project. The goal of the project was to create an open-access biomaterials database through automated curation from scientific literature. The solution consists of a custom ontology and an orchestrated data pipeline for ingestion, annotation and storage/search. This article will touch upon all the components, with a focus on the development and utilization of the ontology.
Ontology
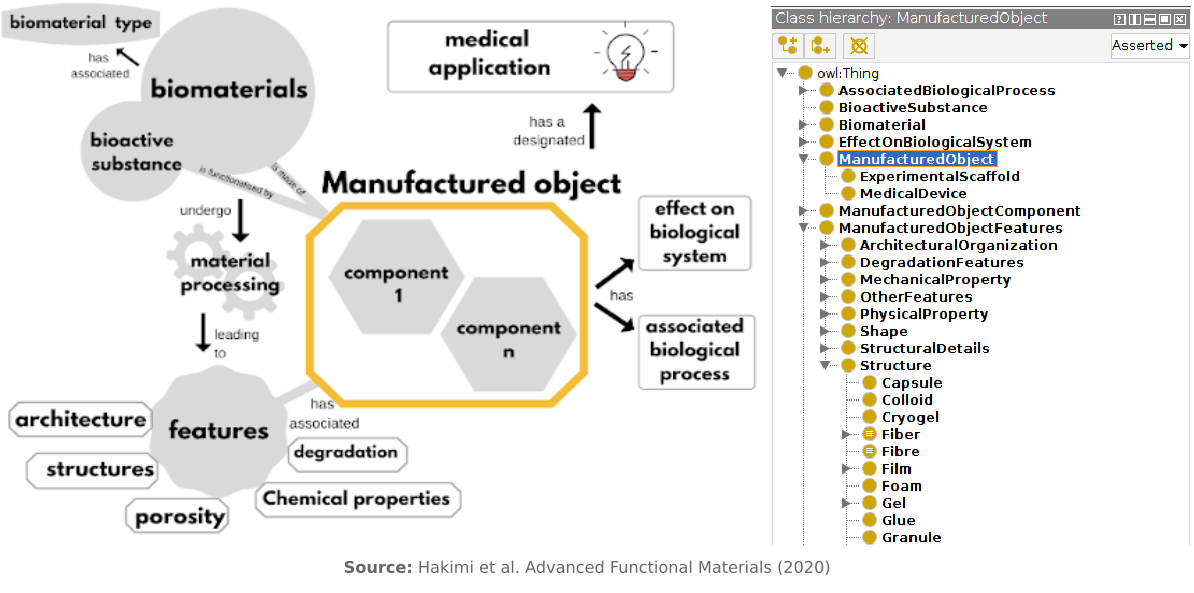
The custom ontology, called Devices, Experimental scaffolds and Biomaterials Ontology (DEB), was designed to categorize scientific information related to biomaterials. The need arose from the interdisciplinary nature of biomaterials research, which causes a lack of standardized taxonomy and resource indexing. To cope with the complexity of the subject and best serve the purpose of literature curation, the DEB ontology was created using a hybrid approach that leveraged both expert knowledge and text-mining techniques:
- The top-down phase involves manual definition of upper-level classes by domain experts.
- The bottom-up phase employed text-mining to automatically populate lower-level concepts and terms, based on a gold-standard set of relevant abstracts retrieved from PubMed.
- Multiple top-down and bottom-up iterations were performed to refine the ontology structure for coherence.
- For interoperability, all relevant terms were cross-referenced with other public ontologies/taxonomies such as OBO and NCIT.
- Finally, the approach, the gold standard set and the ontology were peer reviewed by a global network of biomaterials researchers through survey.
Data Ingestion
The primary data source for the DEBBIE database is PubMed. Using PubMed APIs, abstracts of the most recent scientific publications were retrieved and standardized using an automated pipeline. Additionally, the pipeline includes a filtration step to remove irrelevant abstracts, based on a Support Vector Machine (SVM) classifier.
Data Annotation
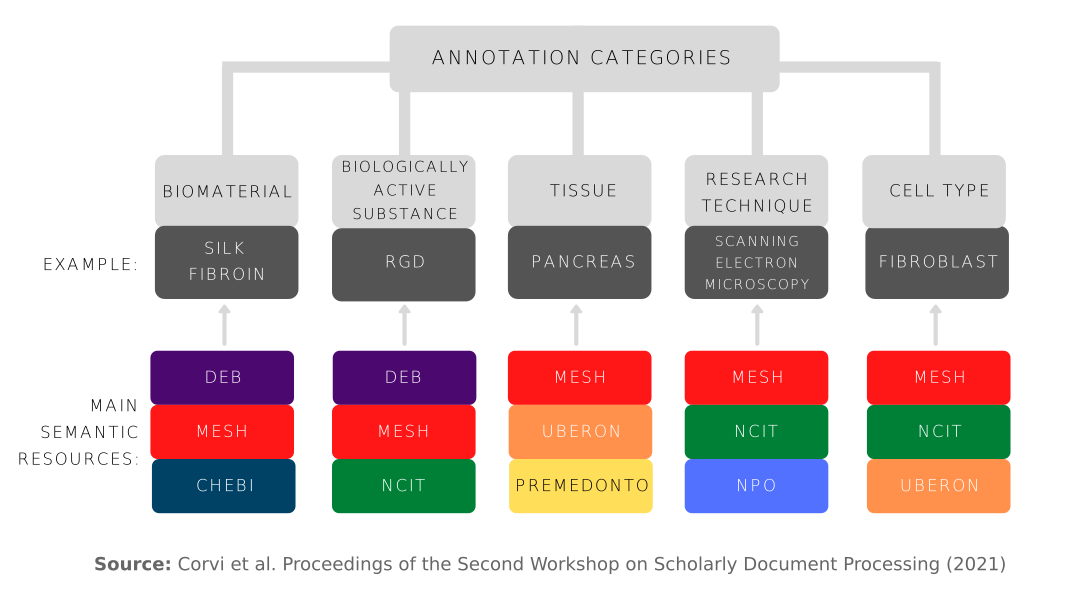
The retrieved PubMed abstracts were annotated using a schema consisting of 17 categories. Each category was mapped to a number of public ontologies/taxonomies including DEB. As machine learning-based approaches were deemed infeasible, the ontologies/taxonomies were converted into dictionaries and then used for lexical-based entity matching. A rule-based post-processing step was also implemented, based on an annotated corpus, to ensure quality control. Finally, all the annotated contents were reviewed and verified by domain experts prior to release for use.
Searchable Database
The annotated PubMed abstracts were converted into JSON and stored in a Mongo database.
At the moment, only a prototype search engine is available for public preview, which has several limitations, such as the search is limited to using a finite set of ~40,000 keywords. For each search session (e.g. “hydrogel”), a summary page with aggregated statistics and a detailed result page are available. The same data can also be accessed through REST APIs.
![DEBBIE User Interface]](/assets/screenshots/2023-02-12-12-28-16.png)
Technical Stack
| Component | Software |
|---|---|
| Ontology editor | Protégé |
| Ontology text-mining | Python NLTK |
| Named entity recognition | Stanford Open NLP |
| NLP Annotation | GATE |
| Data storage | MongoDB |
| Data visualization | ChartJS |
| Packaging & distribution | Docker |
| Task orchestration | NextFlow |
| CI/CD | GitLab |
- BiomaterialsOntologyScientific Software